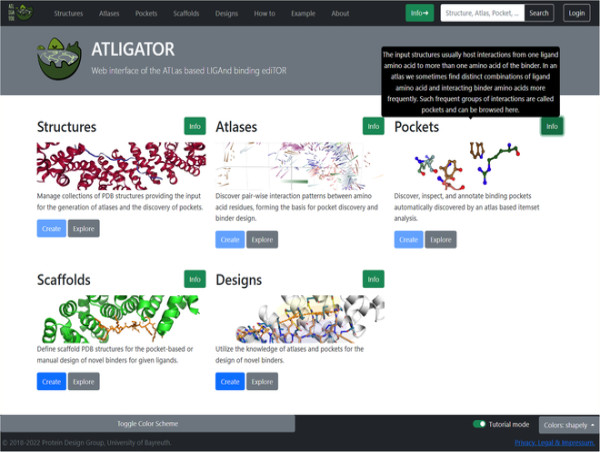
The ATLIGATOR web brings powerful protein design tools to users via a graphical web interface.
CHINA, December 11, 2023 /EINPresswire.com/ -- Proteins can interact with various other proteins and peptides but engineering specific protein–protein interactions is still a challenging task. Researchers at the University of Bayreuth, Germany, however, have made a breakthrough here. They recently integrated the ATLIGATOR software package, which has protein design tools, into a web platform. The interface makes the visually demanding aspect of protein design user-friendly and more accessible.
Can protein design be made more intuitive? The description of protein–protein interactions remains complex and is not an exact science. However, researchers at the University of Bayreuth are hopeful the recent release of ATLIGATOR web—a graphical web interface for the ATLIGATOR software package—will be a game-changer in the field of protein engineering.
Amino acids form the fundamental links in protein–protein or protein–peptide interactions, and some amino acid interactions are more important than others, as they contribute to binding affinity and specificity. One way to study amino acid interactions is to look at them pairwise, assessing their functional groups or energetic yields. An alternative is to profile the groups of amino acids that bind to single residues and understand their interaction patterns. The ATLIGATOR software identifies patterns in frequent pairwise residues using data extracted from complex protein structures.
Taking this a notch ahead, researchers from the Protein Design working group developed the ATLIGATOR web from the ATLIGATOR python package. The web application is more user-friendly as it has a graphical user interface (GUI). It does away with working through the command line—where code and scripting execute functions and extract information—and users can instantly use the tools embedded in the software. An overview of the web platform was published in Volume 5 of BioDesign Research on 4th May 2023. “We wanted to open access to the software by circumventing the need for a local installation. The new GUI visually connects the ATLIGATOR suite of analysis tools and even extends the design capability of the software,” says Professor Birte Höcker, who helms the Protein Design group.
ATLIGATOR web integrates three pages (Structures, Atlases, and Pockets) to analyze binding interfaces, and two pages (Scaffolds and Designs) to design protein interaction sites. The Structures page takes an input protein scaffold and aggregates known structures from similar proteins with a Structural Classification of Proteins—an extended database identifier. The information from this page generates Atlases and Pockets. Pairwise interaction patterns between amino acid residues from the input protein structure undergo identification on the Atlases page. Three-dimensional (3D) plots help explore an amino acid atlas to gain insight into its pairwise interactions. Individual data points on the plots contain comprehensive information about the side chains—via a 3D full-atom view—of the interacting pair. Frequent interactions are then distilled from all data points in Atlases and aggregated on the Pockets page, further helping discover and annotate binding pockets using information gleaned from the amino acid interactions.
Scaffolds and Designs expanded the functionality of ATLIGATOR web. The Scaffolds tool enables users to design protein structures with more than two polypeptides. Here, the ligand chain harbors the amino acid for which the binding pocket is chosen, and the binder is the chain that will be mutated. With Designs, the binder chain can be altered via multiple mutations, and there are two options to mutate the scaffold following the selection of a pocket collection. The first automatically grafts a pocket on the scaffold based on a pocket identified from the pocket collection for each ligand. Since the fit might not be perfect due to geometry constraints, the second option allows the user to customize mutations manually.
Excited by ATLIGATOR web’s potential to democratize the study of protein-peptide interactions, Prof. Höcker concludes, “In this incarnation, ATLIGATOR will empower more users to explore and engineer new binding capabilities by its easy access and the ability to understand the underlying principles.”
DOI
10.34133/bdr.0011
Original Source URL
https://doi.org/10.34133/bdr.0011
Lucy Wang
BioDesign Research
email us here